突出你的新发现:高亮富集结果中的关键通路绘制
今天来学习一篇于2024年12月30日发表在Nature Communications上的文献,标题为:《Integrated multi-omics profiling reveals neutrophil extracellular traps potentiate Aortic dissection progression》。这个文献中的双向富集结果展示了 Scissor+ cells 与 Scissor- cells 差异分析后GO BP富集结果,并高亮了通路中的关键通路:中性粒细胞的激活、趋化和脱颗粒。
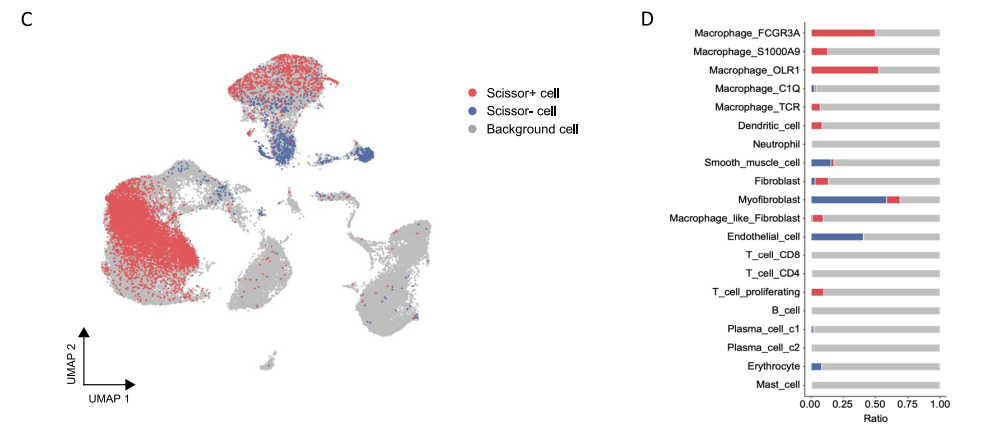
这个文章给我们提供了一种筛选与疾病高度相关的细胞亚群方法,即Scissor算法,它利用单细胞数据和 bulk RNA-seq 数据及表型信息识别与疾病高度相关的细胞亚群。关于什么是 Scissor+ cells 细胞以及 如何鉴定 Scissor+ cells 与 Scissor- cells,我们下期介绍,前面生信技能树也有相关介绍的贴子:
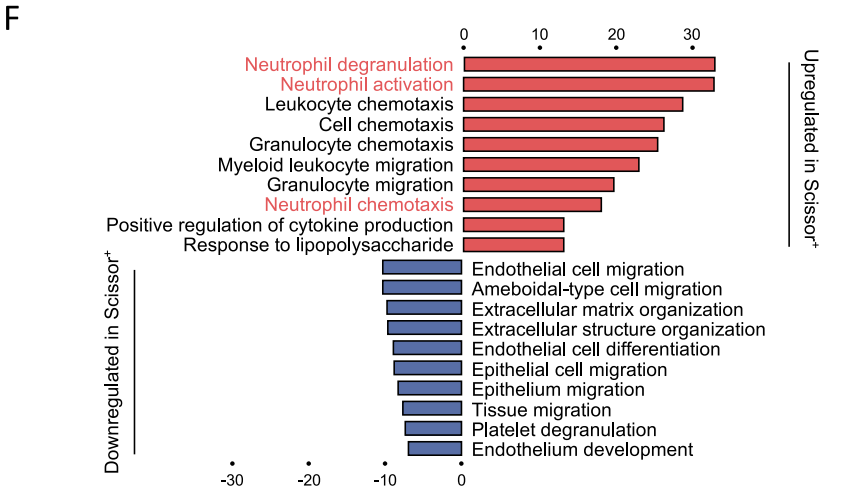
Fig. 2 | A phenotype-associated macrophage subset orchestrates neutrophil within aortic microenvironment. F A bar plot of the GO enrichment of biological processes showing the significantly enriched pathways in Scissor+ cells.
今天先来绘制上面的富集结果双向通路图,并熟悉文章数据背景,分析思路。
数据背景
作者的样本采样以及数据组学思路如下,这里作者还做了预实验蛋白组学:
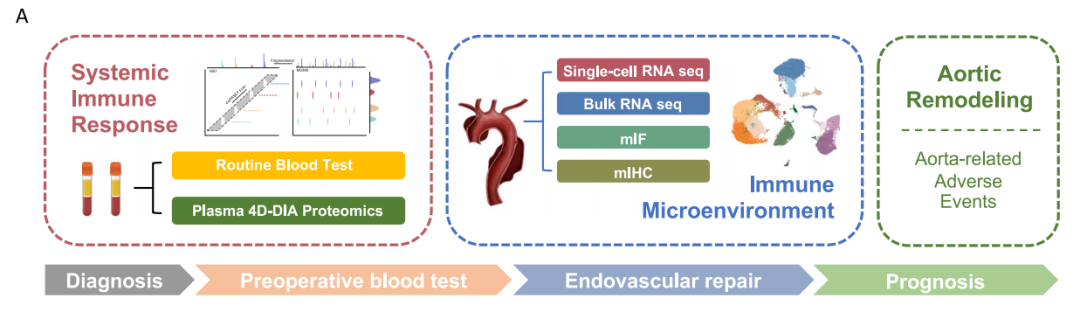
image-20250212184910682
image-20250212184910682
其中单细胞数据来自三名健康个体和三名急性主动脉夹层(AD)患者的主动脉。数据为:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254132。
下载并解压之后的文件如下:
.
├── ATAD1
│ ├── barcodes.tsv.gz
│ ├── features.tsv.gz
│ ├── matrix.mtx.gz
│ └── web_summary.html
├── ATAD2
│ ├── barcodes.tsv.gz
│ ├── features.tsv.gz
│ ├── matrix.mtx.gz
│ └── web_summary.html
├── ATAD3
│ ├── barcodes.tsv.gz
│ ├── features.tsv.gz
│ ├── matrix.mtx.gz
│ └── web_summary.html
├── NA1
│ ├── barcodes.tsv.gz
│ ├── features.tsv.gz
│ ├── matrix.mtx.gz
│ └── web_summary.html
├── NA2
│ ├── barcodes.tsv.gz
│ ├── features.tsv.gz
│ ├── matrix.mtx.gz
│ └── web_summary.html
└── NA3
├── barcodes.tsv.gz
├── features.tsv.gz
└── matrix.mtx.gz
数据简单处理并分析
1、先读取并创建Seurat对象:
###
### Create: Jianming Zeng
### Date: 2023-12-31
### Email: jmzeng1314@163.com
### Blog: http://www.bio-info-trainee.com/
### Forum: http://www.biotrainee.com/thread-1376-1-1.html
### CAFS/SUSTC/Eli Lilly/University of Macau
### Update Log: 2023-12-31 First version
### Update Log: 2024-12-09 by juan zhang (492482942@qq.com)
###
rm(list=ls())
options(stringsAsFactors = F)
library(ggsci)
library(dplyr)
library(future)
library(Seurat)
library(clustree)
library(cowplot)
library(data.table)
library(ggplot2)
library(patchwork)
library(stringr)
library(qs)
library(Matrix)
getwd()
# 创建目录
getwd()
gse <- "GSE254132"
dir.create(gse)
# 下载raw文件夹
# https://ftp.ncbi.nlm.nih.gov/geo/series/GSE163nnn/GSE163558/suppl/GSE163558_RAW.tar
# 使用正则表达式替换:\w{3}$ 匹配每个单词最后的三个字符替换为空字符串,即去掉它们
s <- gsub("(\\w{3}$)", "", gse, perl = TRUE)
s
file <- paste0(gse,"_RAW.tar")
file
url <- paste0("https://ftp.ncbi.nlm.nih.gov/geo/series/",s,"nnn/",gse,"/suppl/",gse,"_RAW.tar")
url
#downloader::download(url, destfile = file)
# 方式一:标准文件夹
###### step1: 导入数据 ######
samples <- list.dirs("GSE254132/", recursive = F, full.names = F)
samples
scRNAlist <- lapply(samples, function(pro){
#pro <- samples[1]
print(pro)
folder <- file.path("GSE254132/", pro)
folder
counts <- Read10X(folder, gene.column = 2)
sce <- CreateSeuratObject(counts, project=pro)
return(sce)
})
names(scRNAlist) <- samples
scRNAlist
# merge
sce.all <- merge(scRNAlist[[1]], y=scRNAlist[-1], add.cell.ids=samples)
sce.all <- JoinLayers(sce.all) # seurat v5
sce.all
# 查看特征
as.data.frame(sce.all@assays$RNA$counts[1:10, 1:2])
head(sce.all@meta.data, 10)
table(sce.all$orig.ident)
library(qs)
qsave(sce.all, file="GSE254132/sce.all.qs")
共得到 53864 个细胞,然后简单质控并降维聚类分群,得到如下注释结果:
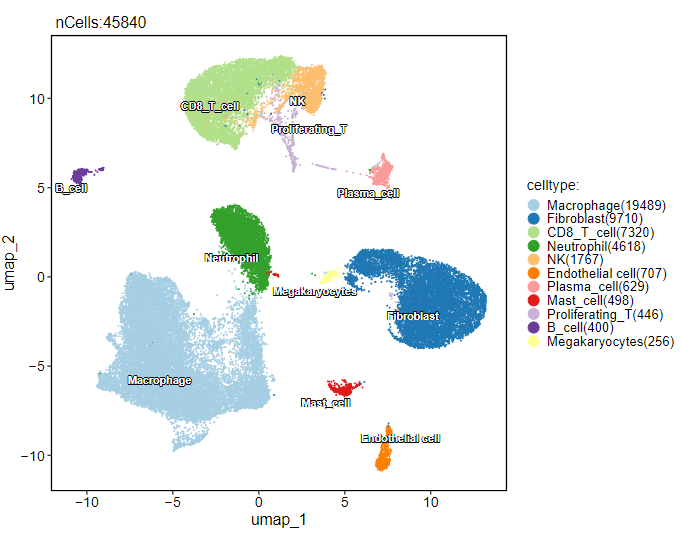
2、功能富集分析
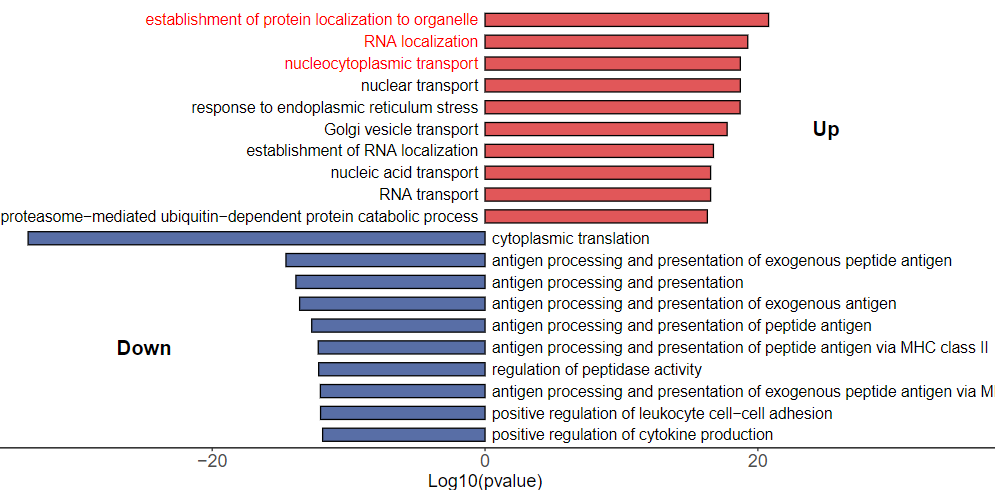
我这里还没有做完 Scissor+ cells 细胞的鉴定,所以本次就随意提取巨噬细胞,分析巨噬细胞在 二分组:健康和急性主动脉夹层(AD)的组间差异并进行功能富集分析,得到一个富集结果用于绘图。
# 加载包
library(org.Hs.eg.db)
library(clusterProfiler)
library(ggplot2)
# 得到 疾病和正常对照组
table(sce.all.int$orig.ident)
sce.all.int$group <- ifelse(grepl("ATAD", sce.all.int$orig.ident), "ATAD", "NA")
table(sce.all.int$group)
table(sce.all.int$group, sce.all.int$orig.ident)
# 提取其中的巨噬细胞
sce_sub <- subset(sce.all.int, idents = c("Macrophage"))
sce_sub
Idents(sce_sub) <- "group"
# 组间差异分析
sce.sub.markers <- FindMarkers(sce_sub, ident.1 = "ATAD", ident.2 = "NA", logfc.threshold = 0.25, min.pct = 0.1)
sce.sub.markers <- sce.sub.markers[sce.sub.markers$p_val_adj < 0.01, ]
sce.sub.markers$name <- rownames(sce.sub.markers)
sce.sub.markers$g <- ifelse(sce.sub.markers$avg_log2FC>0, "up", "down")
head(sce.sub.markers)
table(sce.sub.markers$g)
# 基因转成ENTREZID
ids <- bitr(sce.sub.markers$name, fromType = "SYMBOL",toType = "ENTREZID", OrgDb = "org.Hs.eg.db")
head(ids)
# 合并ENTREZID到obj.markers中
data <- merge(sce.sub.markers, ids, by.x="name", by.y="SYMBOL")
head(data)
gcSample <- split(data$ENTREZID, data$g)
str(gcSample)
## 富集分析
# KEGG
# xx_kegg <- compareCluster(gcSample, fun="enrichKEGG", organism="hsa", pvalueCutoff=1, qvalueCutoff=1)
# GO
xx_go <- compareCluster(gcSample, fun="enrichGO", OrgDb="org.Hs.eg.db", ont="BP", pvalueCutoff=1, qvalueCutoff=1)
res <- xx_go@compareClusterResult
head(res)
## 将富集结果中的 ENTREZID 重新转为 SYMBOL
Symbol <- mapIds(get("org.Hs.eg.db"), keys = sce.sub.markers$name, keytype = "SYMBOL", column="ENTREZID")
head(Symbol)
for (i in 1:dim(res)[1]) {
arr = unlist(strsplit(as.character(res[i,"geneID"]), split="/"))
gene_names = paste(unique(names(Symbol[Symbol %in% arr])), collapse="/")
res[i,"geneID"] = gene_names
}
head(res)

3、绘制
还是大家喜欢的ggplot2定制化绘图,先提取绘图数据并简单处理:高亮其实很简单,一个颜色向量里面设置关注通路的颜色为红色
## 通路筛选Top10
enrich <- res %>%
group_by(Cluster) %>%
top_n(n = 10, wt = -pvalue)
# 绘图数据
dt <- enrich
# 通路排序
dt <- dt[order(dt$Cluster,dt$pvalue, decreasing = F), ]
dt$Description <- factor(dt$Description, levels = rev(unique(dt$Description)))
dt$Cluster <- factor(dt$Cluster, levels = c("up","down"))
# 得到双向横坐标,上调的应该在右边,乘以一个正值
dt$lable <- ifelse(dt$Cluster=="up", 1, -1)
dt$pvalue_loc <- -log10(dt$pvalue)
dt$pvalue_loc <- dt$pvalue_loc * dt$lable
head(dt)
# 调整添加的y轴方向通路的对其方式
dt$lable_hjust <- ifelse(dt$Cluster=="up", 1, 0)
# 对其的x轴起点,上调通路在x轴左边,起点隔0.5, 避免与柱子粘在一起
dt$lable_xloc <- ifelse(dt$Cluster=="up", -0.5, 0.5)
# 设置通路高亮颜色,这里随便选了三条通路进行高亮,标为红色
dt$label_color <- "black"
dt$label_color[1:3] <- "red"
head(dt)
summary(dt$pvalue_loc)
绘图,条形图:
## 绘图
color <- c(up="#e15759", down="#586ea6")
color
p <- ggplot(dt, aes(x = pvalue_loc, y = Description, fill = Cluster)) +
geom_bar(stat = "identity",color="black", width = 0.6) + # 绘制条形图,边框为黑色
scale_fill_manual(values = color) +
scale_x_continuous(limits = c(-34,34)) +
geom_text(data = dt, aes(x=lable_xloc, y = Description, label = Description, hjust = lable_hjust),
color=dt$label_color, size = 4.5) + # 添加通路名称
labs(x="Log10(pvalue)", y=NULL, title="") +
annotate("text", x = 25, y = 15, label = "Up", size=6, fontface = "bold", color="black") +
annotate("text", x = -25, y =5, label = "Down", size=6, fontface = "bold", color="black") +
theme_classic() +
theme( axis.text.x = element_text(size = 15),
axis.title.x = element_text(size = 15),
axis.text.y = element_blank(), # 不显示Y轴标签
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
legend.position = 'none' )
p
ggsave(filename = "Enrich_cometplot.pdf", width = 12, height = 6, plot = p)
结果如下:
看文献时,突然想起导师的日常灵魂发问,深入骨髓那种:这个文章做了什么?如何做的?结论是什么?这个文章你觉得最有意思的地方在哪?
嗯,我觉得文章中如何使用 Scissor算法鉴定与 急性主动脉夹层相关的细胞亚群这个地方比较有意思。这种细胞亚群鉴定出来后,我可以看看他具体是什么细胞类型,他是倾向于一种细胞亚群比如T细胞都是 Scissor+细胞呢还是T细胞里面只有一部分是?如果是后者,他有什么比较特殊的特征吗?这一群细胞可能会发挥什么作用呢?下期,我们来学习!
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-02-12,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

