CUT&Tag 数据处理和分析教程(4)
CUT&Tag 数据处理和分析教程(4)

数据科学工厂
发布于 2025-03-31 19:23:20
发布于 2025-03-31 19:23:20
引言
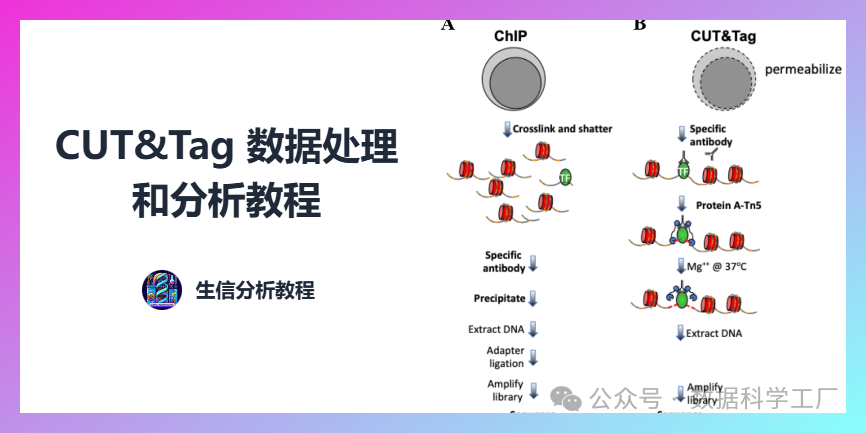
本系列[1] 将开展全新的CUT&Tag 数据处理和分析专栏。想要获取更多教程内容或者生信分析服务可以添加文末的学习交流群或客服QQ:941844452。
报告测序比对总结
对原始读取和唯一比对读取进行总结,以反映比对的效率。高质量数据的比对频率通常应高于 80%。CUT&Tag 数据背景噪声较低,因此在人类基因组中,仅需 100 万比对片段就能为组蛋白修饰提供可靠的分析结果。而对于丰度较低的转录因子和染色质蛋白,下游分析可能需要 10 倍于该数量的比对片段。
我们可以评估以下指标:
- 测序深度
- 比对率
- 可比对片段数量
- 重复率
- 独特文库大小
- 片段大小分布
测序深度
##=== R command ===##
## Path to the project and histone list
projPath = "/fh/fast/gottardo_r/yezheng_working/cuttag/CUTTag_tutorial"
sampleList = c("K27me3_rep1", "K27me3_rep2", "K4me3_rep1", "K4me3_rep2", "IgG_rep1", "IgG_rep2")
histList = c("K27me3", "K4me3", "IgG")
## Collect the alignment results from the bowtie2 alignment summary files
alignResult = c()
for(hist in sampleList){
alignRes = read.table(paste0(projPath, "/alignment/sam/bowtie2_summary/", hist, "_bowtie2.txt"), header = FALSE, fill = TRUE)
alignRate = substr(alignRes$V1[6], 1, nchar(as.character(alignRes$V1[6]))-1)
histInfo = strsplit(hist, "_")[[1]]
alignResult = data.frame(Histone = histInfo[1], Replicate = histInfo[2],
SequencingDepth = alignRes$V1[1] %>% as.character %>% as.numeric,
MappedFragNum_hg38 = alignRes$V1[4] %>% as.character %>% as.numeric + alignRes$V1[5] %>% as.character %>% as.numeric,
AlignmentRate_hg38 = alignRate %>% as.numeric) %>% rbind(alignResult, .)
}
alignResult$Histone = factor(alignResult$Histone, levels = histList)
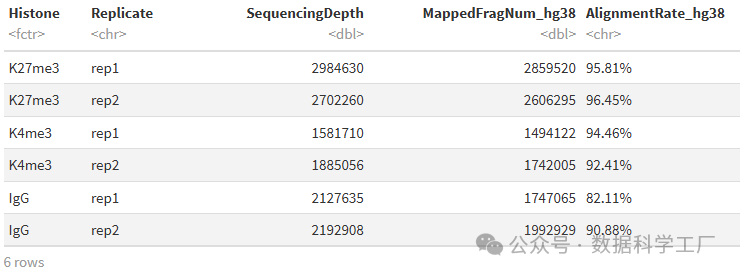
alignResult %>% mutate(AlignmentRate_hg38 = paste0(AlignmentRate_hg38, "%"))
Spike-in alignment
##=== R command ===##
spikeAlign = c()
for(hist in sampleList){
spikeRes = read.table(paste0(projPath, "/alignment/sam/bowtie2_summary/", hist, "_bowtie2_spikeIn.txt"), header = FALSE, fill = TRUE)
alignRate = substr(spikeRes$V1[6], 1, nchar(as.character(spikeRes$V1[6]))-1)
histInfo = strsplit(hist, "_")[[1]]
spikeAlign = data.frame(Histone = histInfo[1], Replicate = histInfo[2],
SequencingDepth = spikeRes$V1[1] %>% as.character %>% as.numeric,
MappedFragNum_spikeIn = spikeRes$V1[4] %>% as.character %>% as.numeric + spikeRes$V1[5] %>% as.character %>% as.numeric,
AlignmentRate_spikeIn = alignRate %>% as.numeric) %>% rbind(spikeAlign, .)
}
spikeAlign$Histone = factor(spikeAlign$Histone, levels = histList)
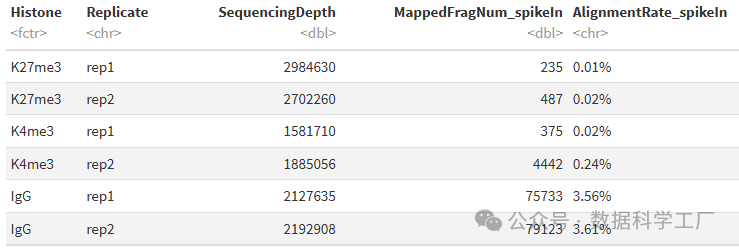
spikeAlign %>% mutate(AlignmentRate_spikeIn = paste0(AlignmentRate_spikeIn, "%"))
比对总结
##=== R command ===##
alignSummary = left_join(alignResult, spikeAlign, by = c("Histone", "Replicate", "SequencingDepth")) %>%
mutate(AlignmentRate_hg38 = paste0(AlignmentRate_hg38, "%"),
AlignmentRate_spikeIn = paste0(AlignmentRate_spikeIn, "%"))
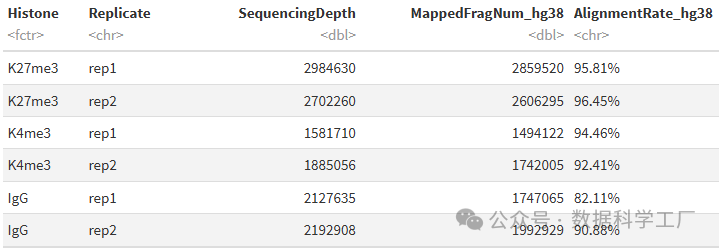
alignSummary
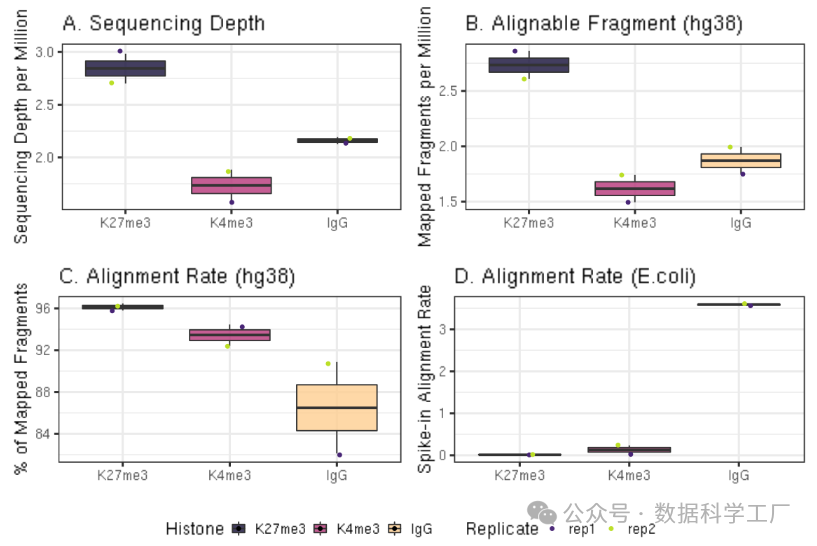
可视化测序深度和比对结果
##=== R command ===##
## Generate sequencing depth boxplot
fig3A = alignResult %>% ggplot(aes(x = Histone, y = SequencingDepth/1000000, fill = Histone)) +
geom_boxplot() +
geom_jitter(aes(color = Replicate), position = position_jitter(0.15)) +
scale_fill_viridis(discrete = TRUE, begin = 0.1, end = 0.9, option = "magma", alpha = 0.8) +
scale_color_viridis(discrete = TRUE, begin = 0.1, end = 0.9) +
theme_bw(base_size = 18) +
ylab("Sequencing Depth per Million") +
xlab("") +
ggtitle("A. Sequencing Depth")
fig3B = alignResult %>% ggplot(aes(x = Histone, y = MappedFragNum_hg38/1000000, fill = Histone)) +
geom_boxplot() +
geom_jitter(aes(color = Replicate), position = position_jitter(0.15)) +
scale_fill_viridis(discrete = TRUE, begin = 0.1, end = 0.9, option = "magma", alpha = 0.8) +
scale_color_viridis(discrete = TRUE, begin = 0.1, end = 0.9) +
theme_bw(base_size = 18) +
ylab("Mapped Fragments per Million") +
xlab("") +
ggtitle("B. Alignable Fragment (hg38)")
fig3C = alignResult %>% ggplot(aes(x = Histone, y = AlignmentRate_hg38, fill = Histone)) +
geom_boxplot() +
geom_jitter(aes(color = Replicate), position = position_jitter(0.15)) +
scale_fill_viridis(discrete = TRUE, begin = 0.1, end = 0.9, option = "magma", alpha = 0.8) +
scale_color_viridis(discrete = TRUE, begin = 0.1, end = 0.9) +
theme_bw(base_size = 18) +
ylab("% of Mapped Fragments") +
xlab("") +
ggtitle("C. Alignment Rate (hg38)")
fig3D = spikeAlign %>% ggplot(aes(x = Histone, y = AlignmentRate_spikeIn, fill = Histone)) +
geom_boxplot() +
geom_jitter(aes(color = Replicate), position = position_jitter(0.15)) +
scale_fill_viridis(discrete = TRUE, begin = 0.1, end = 0.9, option = "magma", alpha = 0.8) +
scale_color_viridis(discrete = TRUE, begin = 0.1, end = 0.9) +
theme_bw(base_size = 18) +
ylab("Spike-in Alignment Rate") +
xlab("") +
ggtitle("D. Alignment Rate (E.coli)")
ggarrange(fig3A, fig3B, fig3C, fig3D, ncol = 2, nrow=2, common.legend = TRUE, legend="bottom")
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-03-30,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

