CUT&Tag 数据处理和分析教程(2)
CUT&Tag 数据处理和分析教程(2)

数据科学工厂
发布于 2025-03-06 21:58:42
发布于 2025-03-06 21:58:42
FastQC 质量控制
- 下载FastQC
##== linux command ==##
mkdir -p $projPath/tools
wget -P $projPath/tools https://www.bioinformatics.babraham.ac.uk/projects/fastqc/fastqc_v0.11.9.zip
cd $projPath/tools
unzip fastqc_v0.11.9.zip
- 运行fastQC以进行质量检查
##== linux command ==##
mkdir -p ${projPath}/fastqFileQC/${histName}
$projPath/tools/FastQC/fastqc -o ${projPath}/fastqFileQC/${histName} -f fastq ${projPath}/fastq/${histName}_R1.fastq.gz
$projPath/tools/FastQC/fastqc -o ${projPath}/fastqFileQC/${histName} -f fastq ${projPath}/fastq/${histName}_R2.fastq.gz
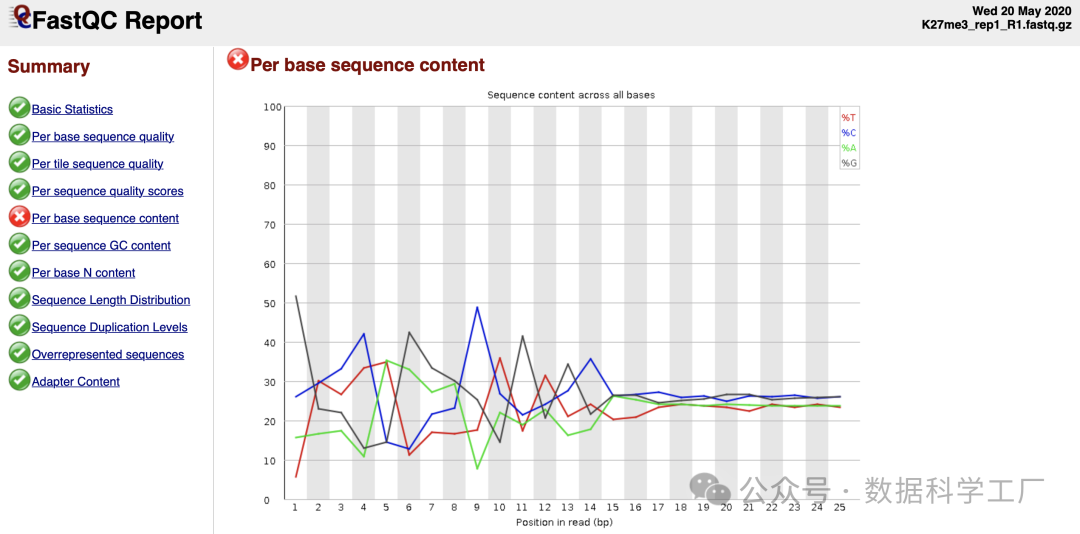
- 质量检查结果
质量检查参考:https://www.bioinformatics.babraham.ac.uk/projects/fastqc/bad_secorence_fastqc.html
合并重复
有时候,为了提高效率,样本会被分成几个泳道来测序,通常在比对前会把它们混合在一起。如果你想看看同一个样本在不同泳道测出来的序列是否一致,可以直接跳过这一步,分别对每个测序文件( fastq 文件)进行比对。
##== linux command ==##
histName="K27me3_rep1"
mkdir -p ${projPath}/fastq
cat ${projPath}/data/${histName}/*_R1_*.fastq.gz >${projPath}/fastq/${histName}_R1.fastq.gz
cat ${projPath}/data/${histName}/*_R2_*.fastq.gz >${projPath}/fastq/${histName}_R2.fastq.gz
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-03-04,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

